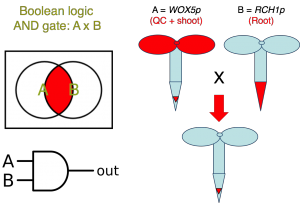
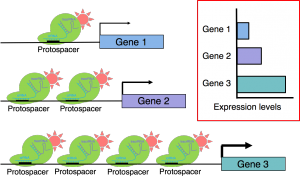
Biology research is often restricted by the choice of promoters available to drive the expression of genes of interest in complex patterns and at desired levels. To overcome this limitation, we are developing a series of easily programmable CRISPR-based synthetic genetic circuits capable of integrating multiple inputs from previously described synthetic and natural promoters to produce novel expression patterns. The new tools take advantage of crRNA:tracrRNA gRNA pairs to combine inputs from two or more different drivers/promoters to delimit where a dCas9-based synthetic transcription factor activates or represses target gene expression. The proposed approach rests on the generation of two or more pseudo-orthogonal gRNA pairs and is compatible with the construction of all basic logic gates to produce multiple derived patterns of expression. In theory, this technology can produce hundreds of orthogonal genetic components that in combination can perform complex logic operations. The new strategy is not only scalable but also enables tuning of the output gene expression levels, thus providing the unprecedented degree of flexibility. To demonstrate the utility of the new approach, we will trigger local auxin production in specific root tissues of an auxin-deficient mutant to control root architecture.