What is recombineering?
What is recombineering?
Recombineering is a homologous-recombination-based technology that allows, among other things, for the insertion of a tag [such as GFP] into the gene of interest contained in a bacterial artificial chromosome (see cartoon). This construct can then be transferred to the plant genome to study, for example, the expression pattern of the gene of interest. To find TAC or BAC clones containing the genes of interest in Arabidopsis and tomato you can use our genome browsers (Araport11 TACs or Tomato2.5 to Araport11). You can also find our recommended TAC clones and primers for a typical N-terminus or C-terminus tagging experiment in the “Recombineering track” of our Araport11 TACs genome browser. You can also use our MatLab GUI primer design tool to customize your recombineering experiment.
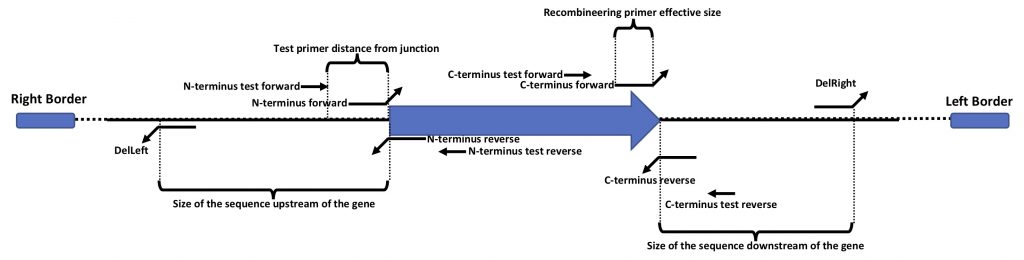
This is a graphic description of the terminology we use for the primer design.
What are the main advantages of using recombineering?
There are two main advantages of using this system:
1) Size does not matter. Small or big genes can be tagged at any position.
2) The expression patterns obtained reflect more accurately those of the endogenous genes. By using large TAC clones (transformation-ready bacterial artificial chromosomes), the gene of interest along with tens of thousands of base pairs of flanking genomic DNA can be included in the recombineering construct. In other words, all regulatory sequences of a gene could be incorporated.
Are there any limitations?
The plant transformation efficiency is significantly lower for large TAC clones than for regular binary vectors. Nevertheless, generation of 15-20 independent lines should not be a problem